 PEPPER in a nutshell
PEPPER in a nutshell
Contents
Introduction
Welcome to the PEPPER-Pipeline project! To get immediately started on collaborating, view development guidelines. We welcome contributors from all backgrounds!
The development of the PEPPER-Pipeline is focused on optimizing an automated, flexible, and easy-to-use preprocessing pipeline dedicated to EEG preprocessing.
Following the optimization of import and preprocessing tools, development will focus on building out a common core of EEG processing tools to handle ERP, time-frequency, and source-based analyses. To find out more about this, view pipeline overview.
Pipeline Overview
The following section details the planning, motivation, and features behind the PEPPER pipeline.
UML Diagrams
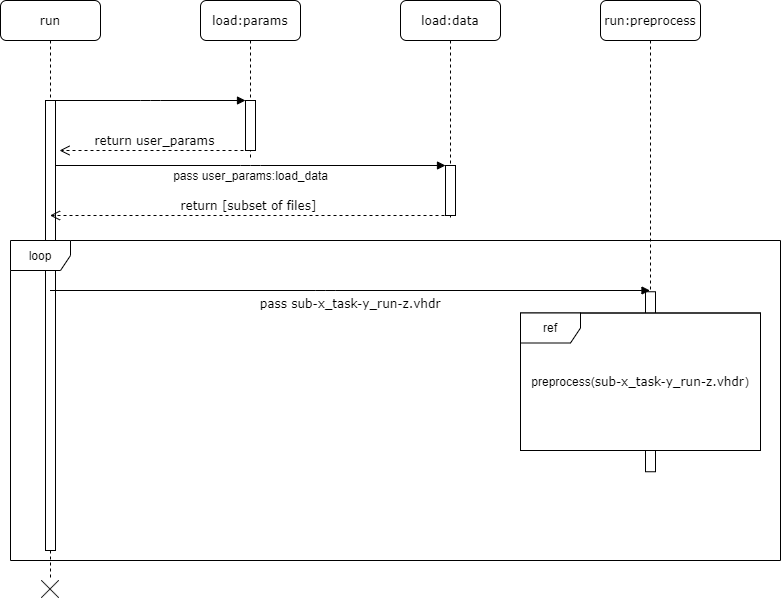
UML diagram for run, which references to run:preprocess
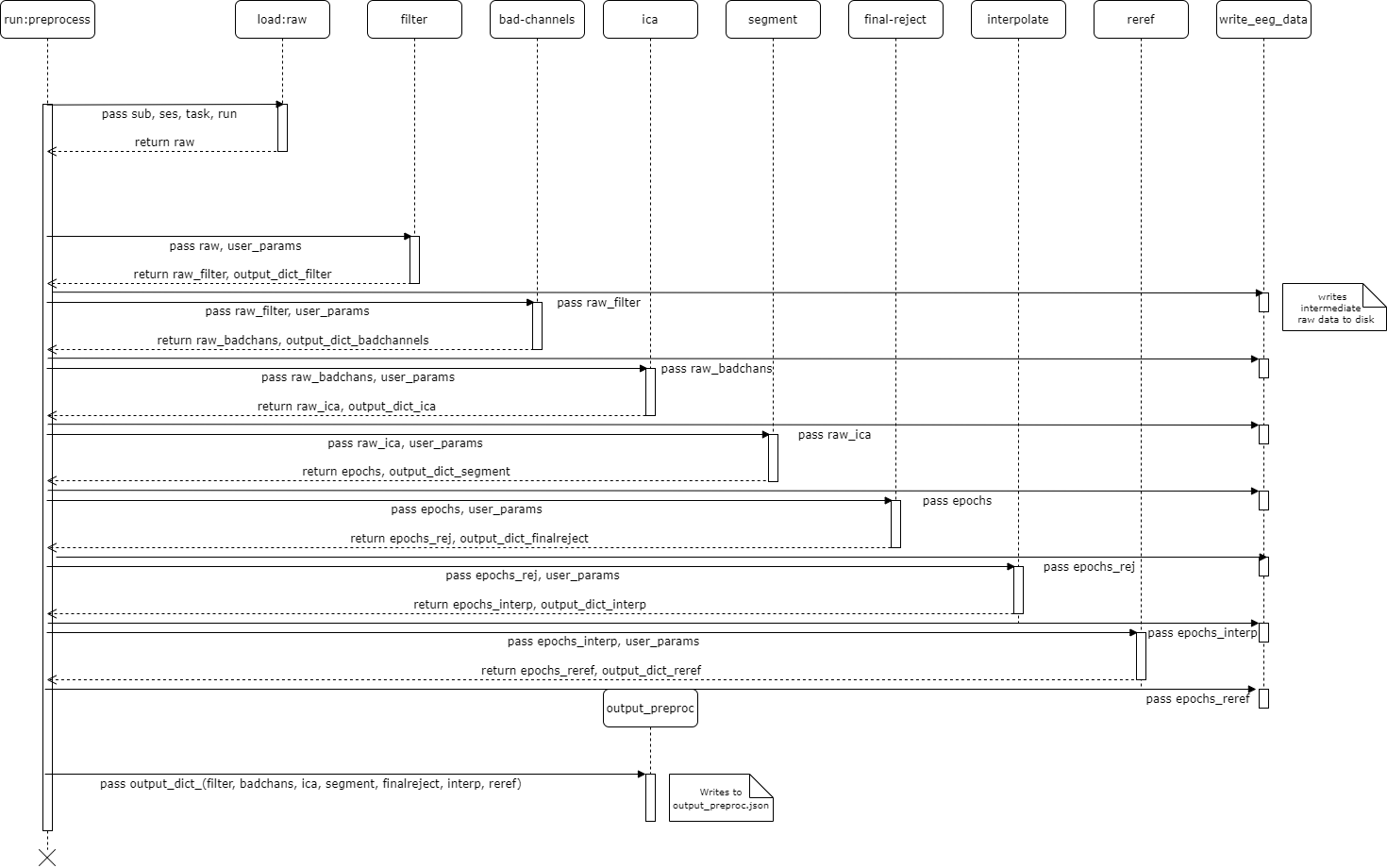
UML diagram for run:preprocess
The UML diagrams above detail the discrete pipeline steps of the default input_config.json file:
-
load:data(pipeline input)A subset of raw data described in
load_datais extracted. -
run:preprocessThe main script calls a series of functions, each one executing a step of the pipeline. All follow the same standard format: each feature always receives an EEG object and unpacked variables from the
paramsdictionary in the main script.Additionally, each pipeline step returns an EEG object and a dictionary describing the changes that occurred to that EEG object.
Motivation behind each pipeline step is described in features.
-
write:outputAt the very last step of the pipeline, each respective output is passed to a
writemodule that transforms the summed outputs into a comprehensive file.
Together, the contents of input_config.json and output_preproc_sub-A_ses-B_task-C_run-D_eeg.json define all details necessary to describe (such as in the methods and results section for a journal publication) the manipulations of the pre-processing pipeline and its outputs.
A long-term goal is to automate the writing of these journal article sections via a script that takes these two files as inputs.
Features
1-Filter
- High pass filter the data using MNE functions
- Read in the “high pass” “low pass” fields from the
user_params.jsonfile to define filter parameters
2-Reject Bad Channels
- Auto-detect and remove bad channels (those that are “noisy” for a majority of the recording)
- Write to output file (field “globalBad_chans”) to indicate which channels were detected as bad
3-Independent Component Analysis
Overview: ICA requires a decent amount of stationarity in the data. This is often violated by raw EEG. One way around this is to first make a copy of the EEG data using automated methods to detect noisy portions of data and then remove these sections. ICA is then run on the copied data after cleaning. The ICA weights produced by the copied dataset are copied back into the original recording. In this way, we do not have to “throw out” sections of noisy data, but we are still able to derive an improved ICA decomposition.
- Prepica
- Make a copy of the EEG recording
- For the copied data: high pass filter at 1 Hz
- For the copied data: segment by epoch to “cut” the continuous EEG recording into arbitrary 1-second epochs
- For the copied data: use automated methods (voltage outlier detection and spectral outlier detection) to detect epochs that are excessively “noisy” for any channel
- For the copied data: reject (remove) the noisy periods of data
- Write to the output file which segments were rejected and based on what metrics
- ICA
- Run ICA on the copied data
- Copy the ICA weights from the copied data back to the pre-copy data
- Rejica
- Use automated methods (TBD) to identify ICA components that reflect artifacts
- Remove the data corresponding to the identified artifacts
- Write to the output file (field “icArtifacts”) which ICA components were identified as artifacts
4-Segment
- Segment by epoch to “cut” the continuous data into epochs of data such that the zero point for each epoch is a given marker of interest
- Write to output file (field “XXX”) which markers were used for epoching purposes, how many of each epoch were created, and how many milliseconds were retained before/after the markers of interest
5-Final Reject Epochs
- Loop through each channel. For a given channel, loop over all epochs for that channel and identify epochs for which that channel, for a given epoch, exceeds either the voltage threshold or spectral threshold. If it exceeds either threshold, reject the channel data for this channel/epoch
- Write to the output file (“field XXX”) which channel/epoch intersections were rejected
6-Interpolate
- Interpolate missing channels, at the epoch level, using a spherical spline interpolation, as implemented in MNE
- Interpolate missing channels, at the global level, using a spherical spline interpolation, as implemented in MNE
- Write to output file (field “XXX”) which channels were interpolated and using what method
7-Re-reference
- Re-reference the data to the average of all electrodes (“average reference”) using the MNE function
- Write to output file (field “XXX”) which data were re-referenced to average
RoadMap
Pre-release
- Minimal, yet complete, pipeline implemented
- Performs all standard preprocessing steps
- Validated, but not optimized
- Containers are stable
- Testing suite is stable
- Standards for community-driven contributions established
- Contributor documentation
- Initial governance structure and credit assignment standards established
Release 0.1
- Modules to allow running in parallel on local or remote (HPC) controlled by the same parameters in the input_params.json file (auto-generates Slurm scripts for HPC)
- Preprint posted to bioRxiv
- Initial data quality assessment suite is stable
- Pipeline meets/exceeds at least one commonly used and published pipeline on data quality metrics
- Initial set of standard input parameters for child, adolescent and young adult data
- Updated functions:
- Filter
- ICA
- Standalone import-feature-io template
- User-end documentation
- Updated/refined governance structure and credit assignment standards
Release 1.0
- Optimized for infant, child, adolescent, and young adult data
- Standard input parameters available
- Revised data output formats to bring in line with emerging BIDS-EEG derivatives
- Expanded data quality assessment suite
- Functions relying on electrode locations updated to use 3-dimensional coordinates based on age-appropriate head model
- Integrated with DataLad
- Verbose .log output files
Release 2.0
- Data quality optimization module
- Module for downloading remote datasets
- Integrated with DataLad
- GUI for generating input_params.json file
- Web-based GUI and automated connection to computational resources
- Expanded set of norms for standard EEG features
Development Guidelines
Identify Issues or Enhancements
If you believe a new issue needs to be added to the list of open issues, feel free to create a new issue and select the appropriate template that suits the indicated change.

Once an issue has been created, the original author can likewise immediately assign themselves and start coding or documenting as described in contribute to the code.
Contribute to the Code
To get started on coding, follow the steps below. Note that you must have a GitHub account to collaborate on this project. All quoted commands are executed in your shell.
-
Fork the repo to your GitHub account by clicking on the “Fork” button on the top right corner of the PEPPER repository:

- Clone the repository to your local machine, with
git clonein your terminal. Be sure to replaceuserbelow with your own GitHub username.git clone https://github.com/[user]/pepper-pipeline.git cd pepper-pipeline -
Build and activate a container using the OS-relevant files (see
containers/README.md). - Switch to the branch that you plan to contribute to.
- If work on this issue has already begun, then fetch and checkout the active branch and then create a sub-branch.
cd pepper-pipeline git checkout dev-feature-issue git checkout -b dev-feature-issue-name -
If this issue has not begun development, then create a new branch and then create a sub-branch.
cd pepper-pipeline git checkout -b dev-feature-issue git checkout -b dev-feature-issue-name
-
Implement changes (commit often!).
git add file1 file2 git commit -m "Attached flux capacitor" -
After you complete all your intended commits, push changes to branch.
git push origin dev-feature-issue-name -
Create a pull request using the GitHub GUI.
Containers
Please use the dockerfile & singularity recipe located in containers/. Directions on installation and usage are located in container/README.md.