 Visualization by Yale Center for Research Computing
Visualization by Yale Center for Research Computing
Contents
Connecting
To use the HPC, a lab member must either use:
- on-campus WiFi
- the FIU VPN to access the FIU intranet. Instructions are available here.
Once connected to either on-campus WiFi or the VPN, use Google Chrome to access the site hpcgui.fiu.edu.
Access Methods
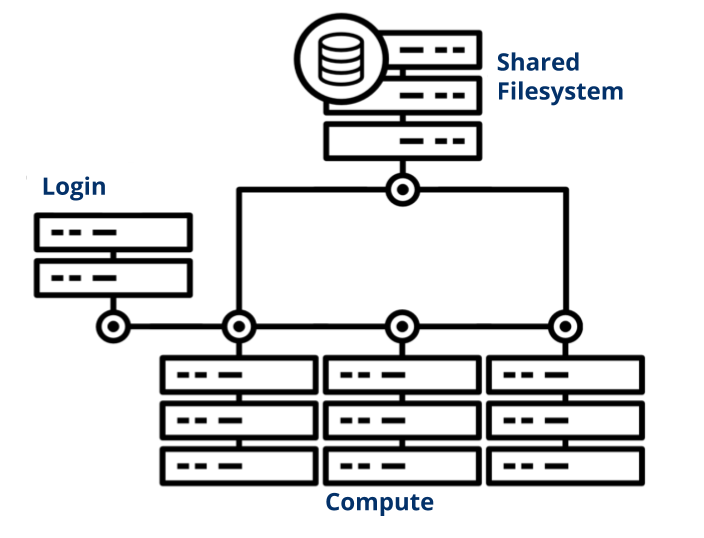
Login Node
The login node, also known as the head node, is the primary HPC entry point for submitting jobs and transferring small amounts of data.
From the HPC GUI login page, select “Clusters,” then “Panther Shell Access;” 
When prompted, input your password. (It will no look like you are typing anything, but persevere and hit Enter!)
A prompt will indicate a successful login:
#######################################################################
Welcome to the FIU Instructional & Research Computing Center (IRCC)
#######################################################################
From here, you can use the command line as you would use any shell. You can also submit batch jobs to the compute-node with the “sbatch” command.
If you are feeling fancy, you can access this directly without logging in via the browser. Simply open your shell and type:
ssh YOURLOGIN@hpclogin01
Or, if accessing from outside FIU HPC:
ssh YOURLOGIN@hpclogin01.fiu.edu
You will be prompted to input your password, and then you can navigate the HPC as you would on the browser. To return to using your shell to access files on your local computer, simply execute an exit command. (Note: this method is called “secure shell (SSH), which comes pre-installed on Windows10 and MacOS. Older Windows versions are available for install here.)
Visual Node
The visualization node is used for directly editing files on the cluster and for GUI manipulation. There are two options:
- the file navigator
- an interactive desktop
The file navigator is similar to a Windows Explorer or Mac Finder type interface. It is useful for viewing folder structure and contents from a high level.
If more interactive access is required, for instance downloading Pavlovia data and uploading to the HPC, this can be accomplished on the interactive desktop. The desktop is resource intensive, so it is best reserved for the kinds of operations that cannot be accomplished via the shell or file navigator. And, because of the relationship with GitHub (see below), it is imperative that files (excluding those in sourcedata/) are never added, deleted, or modified via the interactive desktop.
Structure
The NDCLab follows the structure listed below. 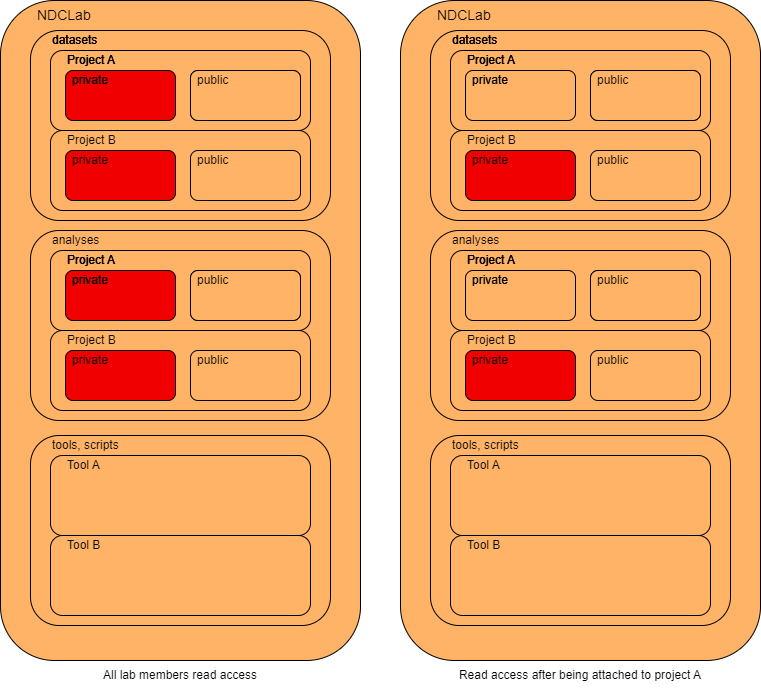
The left and right diagrams represent the read, write, and execute permissions for a general lab member and a lab member attached to a given project, respectively. Each color corresponds to the following group:

The default path to the labs’ folder on the HPC is /home/data/NDClab/
All lab members have general read access to the labs’ projects, including datasets, analyses, and tools, but only a select few members are granted read access to specific data folders or granted write access on specific project. This results in specific privileges given to select members to ensure data compliance. For example, all lab members have read and execute privileges for any public data on the cluster, but only “Project-A” lab members will be able to read and execute the private section of project-A/. Write permissions to a project is handled on a case-by-case basis depending on the lab member’s role in the project.
The main directories – datasets, tools, and analyses – are described below.
datasets
Data collection projects have a directory in this folder to house de-identified and encrypted data. Project leads, approved project members, and lab staff have read and write access to the entire directory, while lab members who are not involved in the project are able to view publicly-available data.
In addition, publicly-available datasets may be replicated to their own directories for additional analyses to be performed by lab members.
tools
This folder contains various scripts and software utilized within the lab for organization, data monitoring, and compliance checking.
analyses
Analysis projects have a directory in this folder to house analysis scripts and their derivatives, such as statistics, plots, and completed products.
Relationship with GitHub
Wherever possible, the NDCLab endeavors to operate as a fully open lab, providing public access to our ongoing projects. For this reason, all dataset and analysis directories on the HPC are, in fact, mirrors of a GitHub repository.
Information that is pushed to the main branch of the GitHub repository is mirrored to the associated HPC folder at 1 am EST nightly. This connection is one-way: that is, for the purposes of data security, data on the HPC is not mirrored back to GitHub. In addition, the sourcedata and derivatives folders of dataset directories/repositories are explicitly ignored by Git.
If an immediate sync of an HPC directory to the GitHub remote is required, this can be achieved with a git pull from the HPC shell.
Any changes made to files directly on the HPC (excluding the uploading of data in sourcedata/), will prevent this sync from occurring unless the change is immediately pushed back to the GitHub remote with a git push from the HPC shell.